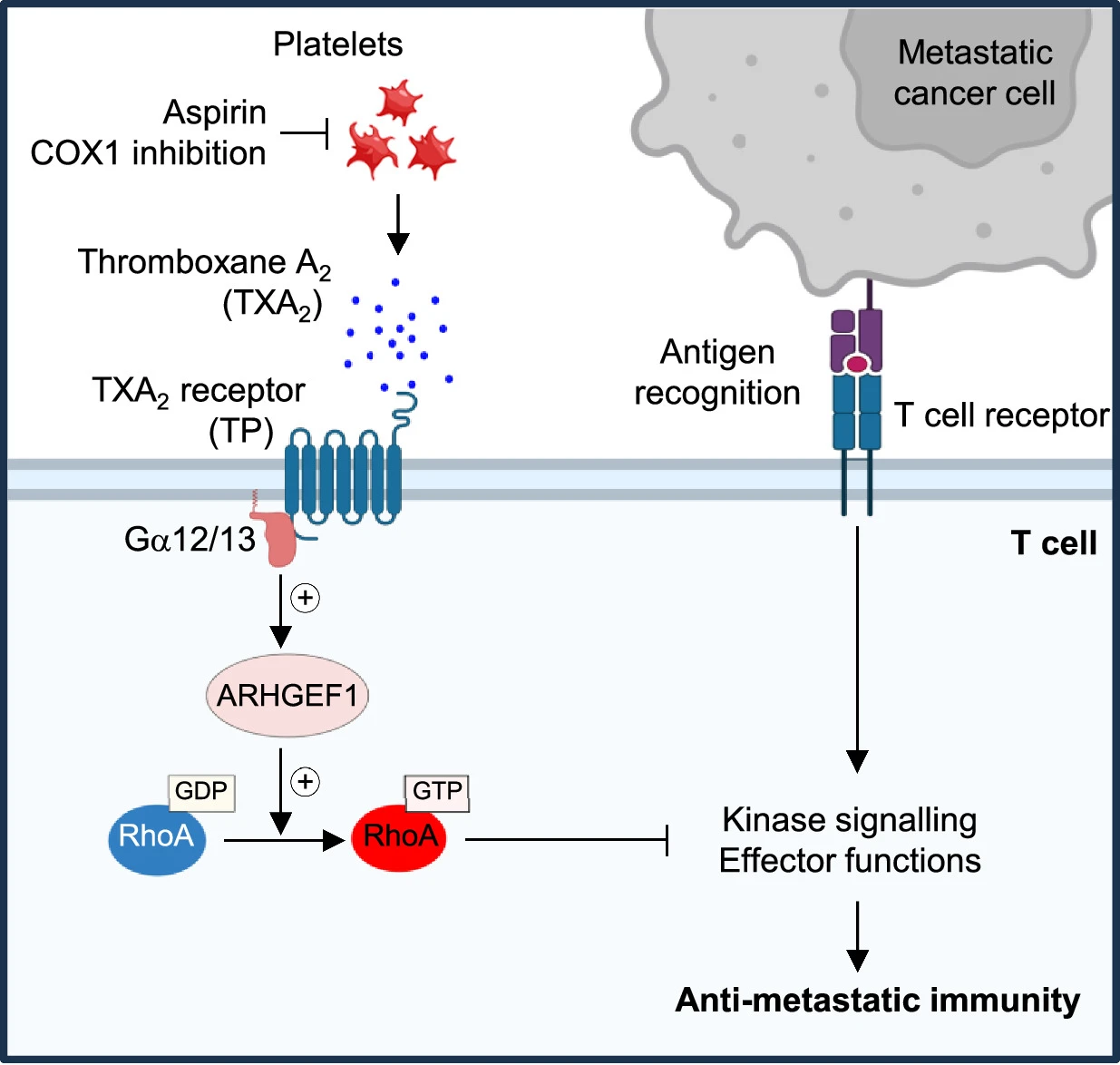
Aspirin prevents metastasis by limiting platelet TXA2 suppression of T cell immunity
아스피린은 T 세포 면역을 억제하는 혈소판 TXA2 제한을 통해 전이를 방지한다 Abstract 전이는 암세포가 원발성 종양에서 멀리 떨어진 장기로 퍼지는 현상으로, 전 세계적으로 암 사망의 90%의 원인이 된다. 전이하는 암세포는…

아스피린은 T 세포 면역을 억제하는 혈소판 TXA2 제한을 통해 전이를 방지한다 Abstract 전이는 암세포가 원발성 종양에서 멀리 떨어진 장기로 퍼지는 현상으로, 전 세계적으로 암 사망의 90%의 원인이 된다. 전이하는 암세포는…
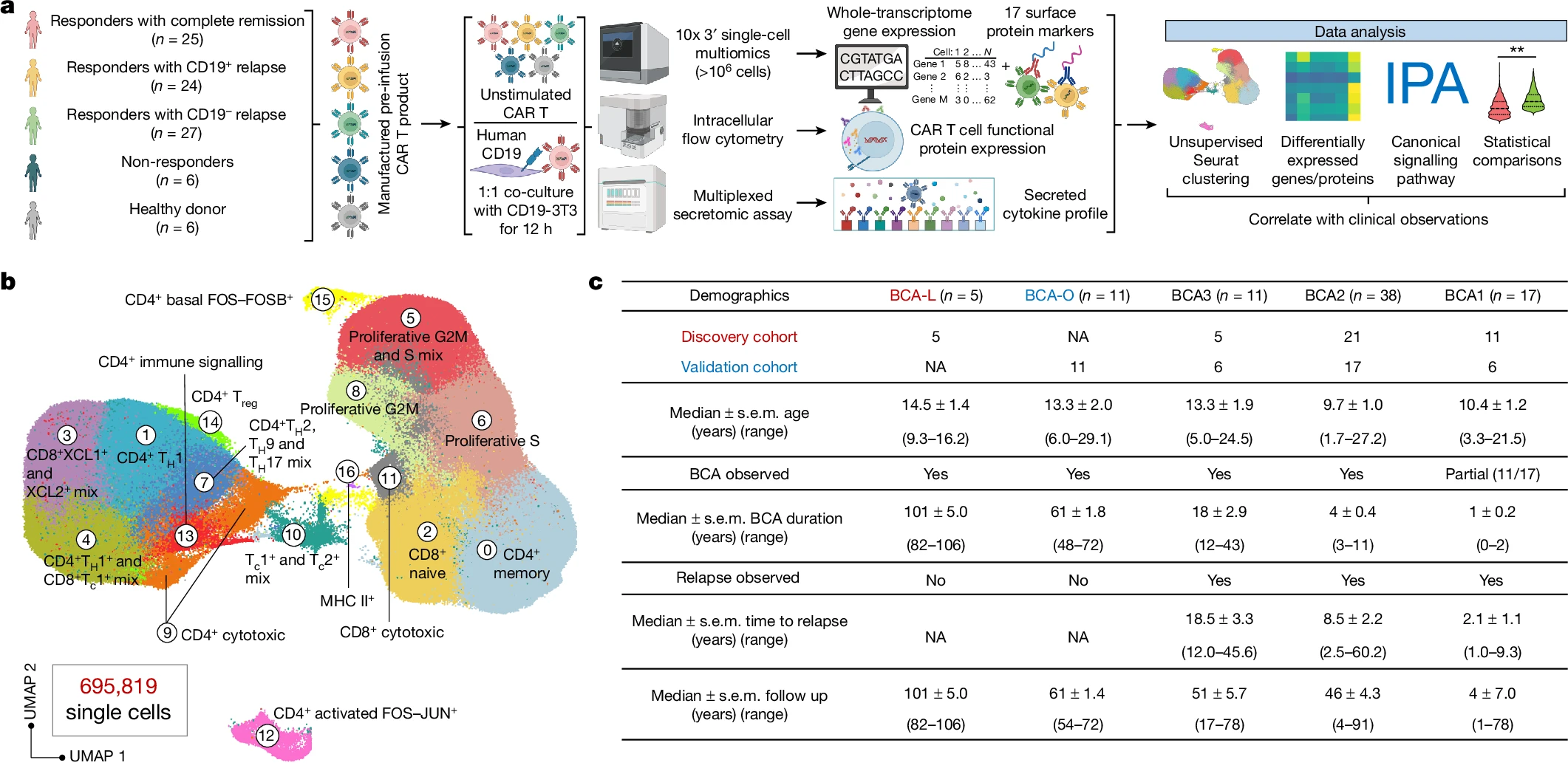
Single-cell CAR T 아틀라스가 8-year leukaemia remission에서 type 2 function을 밝혀낸다 Abstract Acute lymphocytic leukaemia(ALL)에 대한 chimeric antigen receptor (CAR) T cell 치료의 높은 반응률에도 불구하고 약 50%의 환자가 첫해…
Stem cells는 tissue fitness를 유지하기 위해 dead cell clearance를 엄격하게 조절합니다. Abstract 매일 수십억 개의 세포가 우리 몸에서 제거됩니다. Macrophages와 dendritic cells는 죽어가는 세포와 잔해를 engulf하는 역할을 하지만, 많은 epithelial과…
Prime editing의 염색질 맥락-의존적 통제와 후성유전학적 조절 Abstract 이 연구는 prime editing이라는 정밀한 유전체 공학 도구에 Cis-Chromatin 환경이 미치는 영향을 철저히 분석하고자 했습니다. 무작위로 통합된 리포터의 유전체 위치를 mapping하는 고감도 방법을 사용해, 동일한 target 사이트와…
FANCD2–FANCI는 DNA를 감시하고 이중 가닥에서 단일 가닥으로의 접합부를 인식합니다 Abstract DNA 교차결합은 DNA 복제를 차단하며, 이는 Fanconi anaemia 경로에 의해 복구됩니다. FANCD2–FANCI (D2–I) 단백질 복합체는 이 과정에서 중심적인 역할을 하며,…
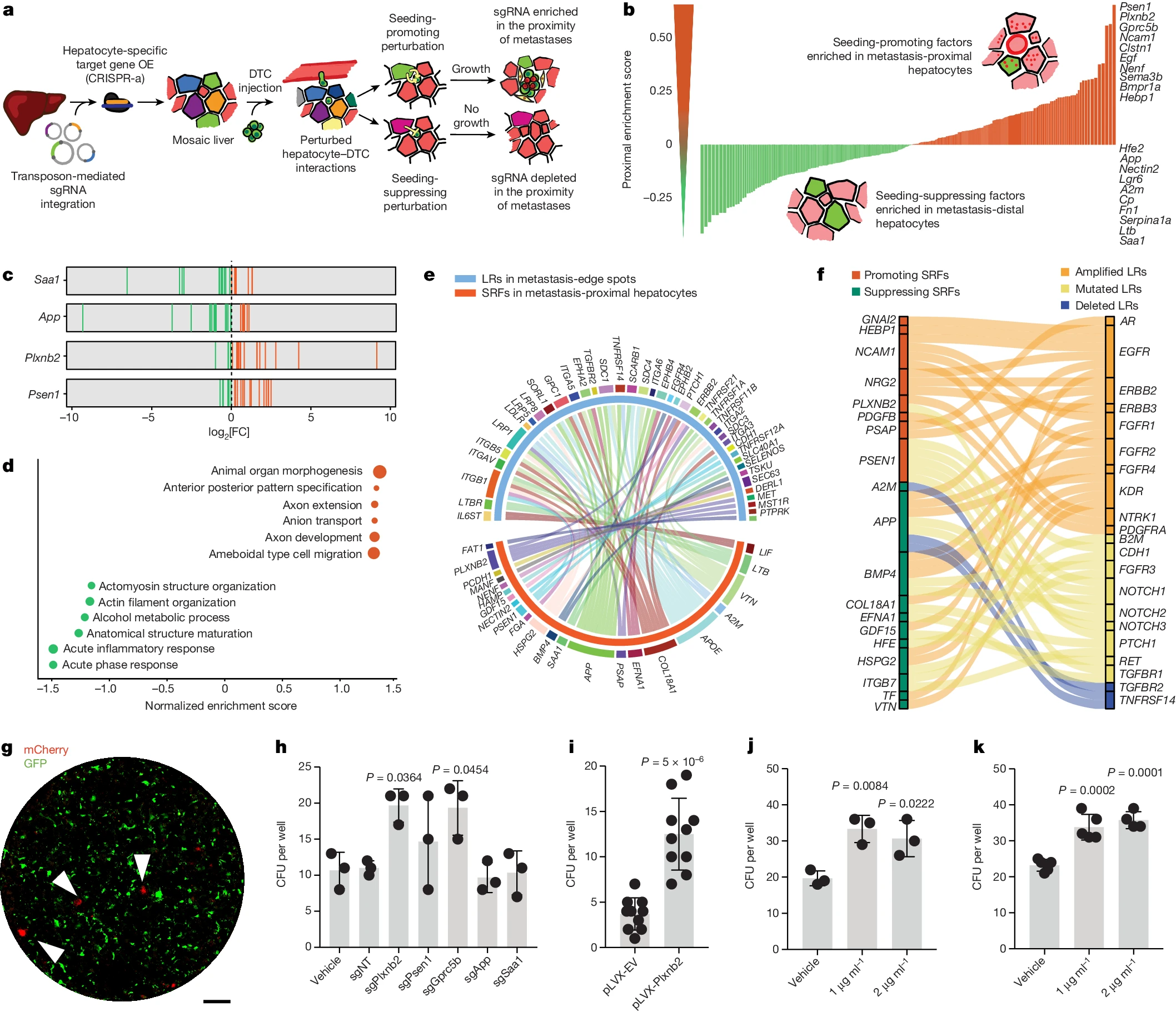
In vivo 상호작용 screening은 전이에 대한 간 유래 constraints를 밝혀낸다 Abstract 전이된 암 세포 중 단 0.02%만이 *명확한 전이(overt metastasis)를 일으킬(seed) 수 있는 것으로 추정된다. 이는 전이의 seeding에 대한 환경적…
TERT의 clonal inactivation는 stem cell competition을 손상시킵니다. Abstract Telomerase는 chromosome ends을 보호하는 nucleoprotein caps인 telomeres를 촉매로 연장하기 때문에 줄기세포 및 암과 밀접한 관련이 있습니다. Telomerase reverse transcriptase (TERT)의 과발현은 telomere와…
dsRNA 형성은 선호적인 nuclear export와 유전자 발현을 유도한다 Abstract mRNA가 핵에서 전사되고 처리된 후, 번역을 위해 세포질로 export됩니다. 이 export는 효모 Saccharomyces cerevisiae에서는 export 수용체 이합체 Mex67–Mtr2에 의해 매개되며, 인간에서는…
Endoplasmic reticulum–plasma membrane contact gradients가 direct cell migration을 유도합니다. Abstract Directed cell migration은 intracellular signaling의 front–back polarization에 의해 주도됩니다. Receptor tyrosine kinases 및 other inputs은 activate local signals를 활성화하여 membrane…
최초의 MKK4억제제는 간 재생을 향진 시키며 간 부전을 예방한다 Abstract 간 질환은 전 세계적으로 연간 200만 명 이상의 사망을 초래하는 대표적인 질환입니다. 급성 혹은 만성으로 손상된 간은 무한한 재생 잠재성을…